Cellular automata experiments
Introduction
I find cellular automata pretty fascinating!
Computation with Automata
I came up with a ruleset that I imagined would work well as a substrate for evolving configurations that model simple functions. It worked pretty well. The search algorithm was that it would score its accuracy with respect to a function and keep or discard random changes in accordance.
It was very fun seeing a machine evolve in front of me pretty rapidly, with a random ruleset I just came up with.
Here’s a configuration that can add one to the binary representation of numbers up to 2^4.
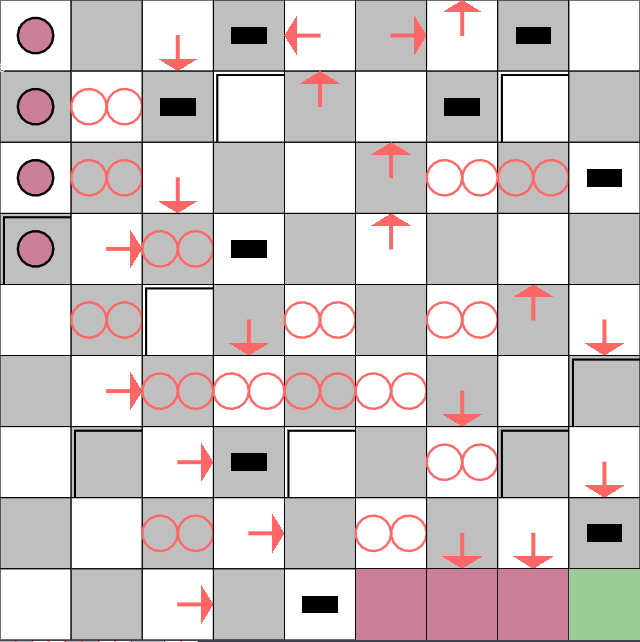
Random Automata
These are some images of automata generated with random rules. I find that around 4 rules is the hotspot for finding a wide range of behaviors.




Persian rug-like

This one actually comes from a bug. I don’t know what the mechanism is, but its result is pretty beautiful - these look like Persian rugs.
Analysis
For a few weeks, I left a program running that generated automata with random rules and scored them across a few qualitative dimensions. The scoring process depended on random initial states and was time expensive, so the scores are pretty inaccurate. However, I did a PCA on the data, which revealed some relationships:
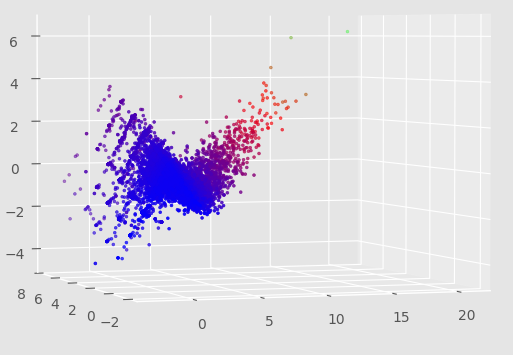
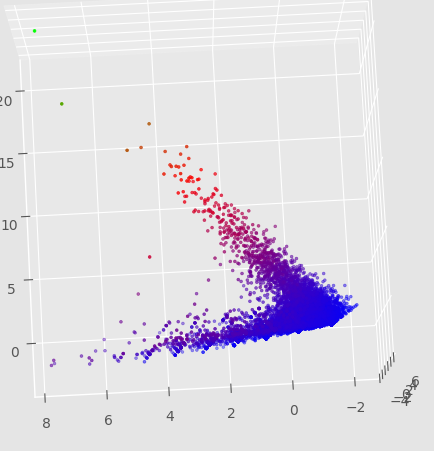
I’m particularly interested in the clusters along certain lines. I wonder if these are a result of the presence of a discrete property with ‘gradations’ - like gliders with a 3-cell seed, then 4-cell seed, etc.
Also, I wonder what those outliers between the two main cluster lines are. Might be as a result of bad data due to randomness, or perhaps those are some pretty interesting automata.
The analysis process:
-- number of repeating exact substructures
add("substructure_count", m.complexity(sim, {iterations=2,x=x,y=y}))
-- number of repeating similar substructures
add("fuzzy_substructure_count", m.complexity(sim, {iterations=2,x=x,y=y,rating_f=function(sim) return #m.repeating_substructures(m.scale_as_img(sim.grid,x/4,y/4),0.7) end}))
-- sensitivity to initial conditions
add("initial_cond_reactivity", m.reactivity(sim, {x=x,y=y,initial_steps=0,wait_steps=20,iterations=5}))
add("stability_reactivity", m.reactivity(sim, {x=x,y=y,initial_steps=20,wait_steps=20,iterations=5}))
-- sensitivity to initial conditions in terms of substructures
add("initial_cond_reactivity_ss", m.reactivity(sim, {x=x,y=y,initial_steps=0,wait_steps=20,iterations=5,comp_states=substructure_compare}))
add("stability_reactivity_ss", m.reactivity(sim, {x=x,y=y,initial_steps=20,wait_steps=20,iterations=5,comp_states=substructure_compare}))